Cheat Sheets
Variable types
R basics
Basic operators + calculation
| Operator | Description | Example |
|---|---|---|
| + | Add | 3 + 4 |
| - | Subtract | 10 - 8 |
| * | Multiply | 6 * 32 |
| / | Divide | 100 / 25 |
| ^ | Exponentiate | 10 ^ 2 |
| == | Equals (logical) | 10 == 10 |
| <- | Assign a value to a name | ten <- 10 |
Check data
| Function | Description | Example |
|---|---|---|
| class() | Check data type | class(iris$Species) |
| ncol() | Check number of columns | ncol(iris) |
| nrow() | Check number of rows | nrow(iris) |
| head() | Return first parts of an object | head(iris) |
| tail() | Return last parts of an object | tail(iris) |
| str() | Check data structure | str(iris) |
| summary() | Generate summary stats | summary(iris) |
| levels() | Return levels of a factor | levels(iris$Species) |
Create data
| Function | Description | Example |
|---|---|---|
| c() | Create a vector | temp <- c("cool", "warm", "hot") |
| factor() | Create a factor | factor(temp) |
Change data type
| Function | Description | Example |
|---|---|---|
| as.numeric | Treat as numeric | as.numeric(temp) |
| as.factor | Treat as factor | as.factor(temp) |
| as.integer | Treat as an integer | as.integer(temp) |
Make simple graphs
| Function | Description | Example |
|---|---|---|
| hist() | Create a histogram | hist(iris$Petal.Width) |
| plot() | Create an x-y plot | plot(iris$Petal.Width ~ iris$Petal.Length) |
| boxplot() | Create a boxplot | boxplot(iris$Petal.Width ~ iris$Species) |
Read + write data
| Function | Description | Example |
|---|---|---|
| read.csv() | Read a .csv file | read.csv("file_name.csv") |
| write.csv() | Write a .csv file | write.csv(iris, "iris.csv") |
| getwd() | Check the working directory | getwd() |
R Markdown
Graph
Grammar of graphics (ggplot2 package)
ggplot(data = penguins, # specify the dataset
mapping = aes(x = flipper_length_mm, # map a variable to the x axis
y = body_mass_g, # map a variable to the y axis
color = species)) + # map a grouping variable to color
geom_point() # choose shapes to represent data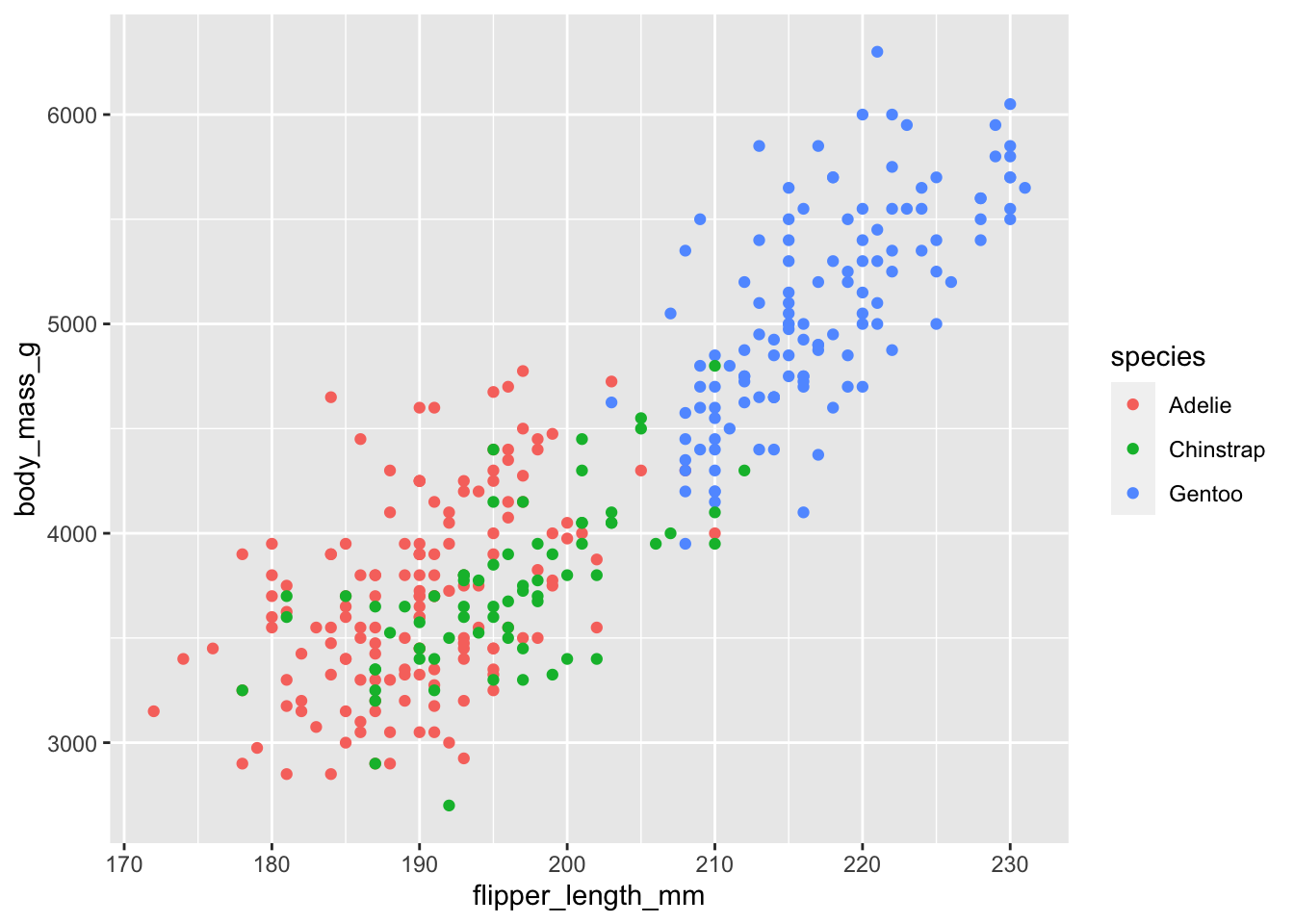
Geom options
| Function | Description |
|---|---|
| geom_col() | Make a barplot (sum) |
| geom_bar() | Make a barplot (frequency/count) |
| geom_boxplot() | Make a boxplot |
| geom_density() | Make a density plot |
| geom_histogram() | Make a histogram |
| geom_point() | Make a scatterplot |
| geom_hline() | Add a horizontal line to a plot (argument
yintercept =) |
| geom_vline() | Add a vertical line to a plot (argument
xintercept =) |
Common modifications
| Function | Description |
|---|---|
| xlim() | Set the range of the x axis (arguments = numeric values for start + end) |
| ylim() | Set the range of the y axis (arguments = numeric values for start + end) |
| xlab() | Set the x axis label (needs to be in quotes) |
| ylab() | Set the y axis label (needs to be in quotes) |
| theme() | Adjust many aspects of graph appearance - fonts, gridlines, legends, etc. |
| theme_bw() | Implement the theme ‘black and white’ |
| theme_classic() | Implement the theme ‘classic’ |
Additional resources
Wrangle
Work with packages
| Function | Description | Example |
|---|---|---|
| install.packages() | Install a package (needed only once) | install.packages("tidyverse") |
| library() | Load a package for use (every session) | library(tidyverse) |
Select + transform data (dplyr package)
| Function | Description |
|---|---|
| filter() | Subset a data frame by condition |
| select() | Select variables in a data frame |
| mutate() | Create new variables |
| arrange() | Order rows in a data frame |
Tidy data (dplyr + tidyr packages)
| Function | Description |
|---|---|
| pivot_longer() | Make a wide dataset long |
| pivot_wider() | Make a long dataset wide |
Additional resources
Summarize
Generate summary statistics (base R)
| Function | Description |
|---|---|
| sum() | Add the values in a vector |
| mean() | Calculate average of values in a vector |
| sd() | Calculate the std deviation of values in a vector |
| length() | Calculate length/# of elements of a vector = sample size |
| sqrt() | Calcualte the square root of a value (needed for std error of the mean) |
Aggregate or summarize by group (dplyr package)
| Function | Description |
|---|---|
| group_by() | Set variable(s) to group a data frame |
| summarize() | Generate summary values by the grouping variable(s)
(e.g. mean = mean(x, na.rm=TRUE)) |
| n() | Calculate the sample size (# of rows) for each group |
t-test
An outline of the steps involved in one-sample t-test:
1. Examine the suitability of data for a t-test
Check variable type to ensure Y is numeric. Helpful functions:
str()
class()
as.numeric() - encodes data as numeric
as.factor() - encodes data as a (categorical) factor
2. Check the normality assumption
Visual inspection
Visualize the distribution of Y
ggplot(data = df, aes(y = Y) + geom_histogram()
OR
hist(df$Y)
Formal test
shapiro.test(df$Y)
- P value < 0.05 means the data are NOT normal
- P value > 0.05 means there is not strong enough evidence to reject the assumption of normality (use your judgement - in combination with the visual inspection)
3. Transform if needed
Two common transformations:
Square root: sqrt(df$Y)
Log base 10: log10(df$Y)
- You can also use
mutatewith these functions to store the transformed variable in the data frame
4. Run the test
Meets normality assumption
Standard t-test example:
t.test(df$Y, alternative = "two.sided", mu = 3, conf.level = 0.95)
- Tests whether the mean of the population of variable Y is significantly different than some value (mu = 3 in this example)
- Two-sided means that it tests both greater and less than the value of mu
- Set to return a 95% confidence interval
Violates the normality assumption
Wilcoxon signed rank test example:
wilcox.test(df$Y, alternative = "two.sided", mu = 3, conf.int = TRUE, conf.level = 0.95)
- Tests whether the median of the population of variable Y is significantly different than some value (mu = 3 in this example)
- Two-sided means that it tests both greater and less than the value of mu
- Set to return a 95% confidence interval
Regression
An outline of the steps involved in linear regression…
See also example code for more detail
1. Assess suitability for linear regression
Are the variables numeric?
Check variable types to ensure X and Y are numeric variables. Helpful functions:
str()
class()
as.numeric()
Are the data points independent?
Remember to ‘check your n’s’ to see how many data points are in the dataset.
What does a row represent?
Is it reasonable to assume that the data points are independent of one another?
What do you know about the experimental design?
Is the relationship linear?
Examine the shape of relationship between X and Y
ggplot(data = df, aes(x = X, y = Y) + geom_point()
OR
plot(df$Y ~ df$X)
What to do if basic requirements are violated:
- Independence
- If it is a borderline case, use visual diagnostics to assess whether related points are clustered (if not, you are probably okay)
- Consider aggregating (e.g. taking a mean across groups) so that you have fewer observations that are independent
- Worst case, you may need to conclude that regression is not appropriate given the experimental design!
- Linearity
- If there simply does not seem to be any relationship, you can still use linear regression and it will likely confirm that there is no relationship between variables.
- If the relationship looks exponential or otherwise curved, it may be possible to transform X, Y, or both to make it linear.
- If there is a more complex curved relationship (e.g. a U shape or hump shape), ideally you would explore non-linear regression methods (e.g. adding an X2 term, called a quadratic model).
2. Fit the model
Using the function lm()…
mod <- lm(Y ~ X, data = df)
3. Check normality and equal variance assumptions
Check for normality + equal variance of residuals:
plot(mod)
Another check for normality of residuals:
hist(mod$residuals)
4. What to do if normality/equal variance assumptions are violated
Option 1: Transform variables to meet assumptions
This is an option when the residuals and/or underlying variables have skewed distributions. The outcomes are easier to interpret for a log transform than a square root transform. It is most common to transform the X variable, but there are situations where it makes sense to transform Y or X and Y.
Natural log: log(df$X)
Log base 10: log10(df$X)
Square root: sqrt(df$X)
- You can also use
mutatefor these operations to store the variable in the dataset
Fit + evaluate the new model on transformed variables… (return to #2 + #3, making sure to fit the model under a new name so you can compare diagnostics/outputs for each)
Option 2: Remove outliers (if outliers are driving the issue)
This is an option if there are a few unusual observations that are far outside the pattern of the rest of the data, and have a heavy influence on fitting the regression line. Outliers will be marked in the residual plots. Their influence on the regression can be assessed using the residuals vs. leverage plot.
Remove outliers (e.g. remove rows # 1, 2, 3)
df_no_out <- df[-c(1,2,3),]
Here is a resource on indexing
Alternatively, you can add a column to your dataset identifying
outliers using mutate and ifelse.
Fit + evaluate a new model if needed… (return to #2 + #3, making sure to fit the model under a new name so you can compare diagnostics/outputs for each)
Option 3: Fit a non-parametric model (Theil-Sen regression)
This is an option if transformation fails to help the data meet assumptions. This method is also not very sensitive to outliers, so it is an alternative to removing them. Keep in mind that this method still assumes linearity and independence.
Using the function mblm()… in the package
mblm
mod <- mblm(Y ~ X, data = df)
Important note!
The function mblm does not work if there are
NA values in X or Y. To solve this problem you can use the
drop_na() function to remove NA values before
fitting the model:
df_no_NA <- df %>% select(X, Y) %>% drop_na()
Then you would fit:
mod <- mblm(Y ~ X, data = df_no_NA)
5. Interpret + graph results
Generate model summary
summary(mod)
Key aspects of the output include:
- The model P value (Is there a linear relationship between X and Y?)
- The adjusted R2 value (How strong is the relationship? How much variation in Y can X explain?)
- The estimates for the intercept and slope (What line best represents the relationship between X and Y? How much does Y change for a 1-unit change in X?)
Resources for interpretation
Make a final plot with a trend line
ggplot(data = df, aes(x = X, y = Y)) +
geom_point() +
Trend line for standard regression with 95% confidence interval around the slope:
geom_smooth(method = "lm", se = TRUE)
Trend line for log10-transformed X with 95% confidence interval around the slope:
geom_smooth(method = "lm", formula = 'y ~ log10(x)', se = TRUE)
Trend line for Theil-Sen regression:
geom_abline(intercept = coef(mod)[1], slope = coef(mod)[2], color = "blue")
ANOVA
An outline of the steps involved in one-way ANOVA…
See also example code for more detail
1. Examine suitability of data for ANOVA
Check variable types to ensure X is categorical and Y is numeric. Helpful functions:
str()
class()
as.numeric() as.factor() - encodes data as a
(categorical) factor
Examine the distributions of X and Y
ggplot(data = df, aes(x = X) + geom_histogram()
OR
hist(df$X)
Examine the distribution of Y by X grouping variable
ggplot(data = df, aes(x = X, y = Y) + geom_boxplot()
OR
boxplot(df$Y ~ df$X)
Note: If you get an error with boxplot, you may have
NA values in your data that are causing problems. At the start of your
code where you are preparing the data, you can remove them using the
function drop_na().
2. Fit the model
Using the function aov()…
mod <- aov(Y ~ X, data = df)
3. Check assumptions
Check for normality + equal variance of residuals:
plot(mod)
Another check for normality of residuals:
hist(mod$residuals)
Another check for equal variance among groups:
boxplot(mod$residuals ~ df$X)
4. Adjust if needed
Consider your options…
- Non-normal residuals: Consider transforming the Y variable
- Unequal variance: Consider Welch’s ANOVA
- Non-normal residuals + unequal variance: Start with transformation, then go from there…
Transform the variable(s)
Log base 10: log10(df$X)
Square root: sqrt(df$X)
- You can also use
mutateto store the outcome of these operations as a new variable
Fit + evaluate (return to #2 + #3, making sure to fit the model under a new name so you can compare diagnostics/outputs for each)
Welch’s ANOVA
Does not assume equal variance.
oneway.test(Y ~ X, data = df)
Removing outliers (last resort)
e.g. remove rows # 1, 2, 3
df_no_out <- df[-c(1,2,3),]
5. Interpret results
Standard ANOVA
Generate model summary:
anova(mod)
If significant differences are present, generate post-hoc comparisons of groups:
HSD.test(mod, "X", group = TRUE, console = TRUE, unbalanced = TRUE)
(in package agricolae)
Note: The argument unbalanced should be
set to TRUE if the sample size is unequal across groups. If
y
Welch’s ANOVA
Generate model summary:
mod
If significant differences are present, generate post-hoc comparisons of groups:
games_howell_test(Y ~ X, data = df)
6. Graph results
Make a final plot with error bars. If you transformed the data, be sure to use the back-transformed means and standard errors.
Spatial data
Key spatial packages
sp: spatial object definitions + some methodssf: will replacespeventually; spatial object definitions + methodsraster: work with raster data, many processing functionsspdep: analysis tools for spatial datatmap: visualization of vector and raster imagesrasterVis: visualization of rater imagesRColorBrewer: helpful package to generate/apply color scales
Resources + guides
sf (spatial object definitions + some methods)
sp (spatial object defintions + some methods, to be
replaced by sf)
tmap (making nice looking maps)
usmap (US political boundaries)
RColorBrewer (create color scales)
Further reading/resources
sessionInfo()R version 4.3.2 (2023-10-31)
Platform: x86_64-apple-darwin20 (64-bit)
Running under: macOS Monterey 12.4
Matrix products: default
BLAS: /Library/Frameworks/R.framework/Versions/4.3-x86_64/Resources/lib/libRblas.0.dylib
LAPACK: /Library/Frameworks/R.framework/Versions/4.3-x86_64/Resources/lib/libRlapack.dylib; LAPACK version 3.11.0
locale:
[1] en_US.UTF-8/en_US.UTF-8/en_US.UTF-8/C/en_US.UTF-8/en_US.UTF-8
time zone: America/New_York
tzcode source: internal
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] palmerpenguins_0.1.1 lubridate_1.9.3 forcats_1.0.0
[4] stringr_1.5.1 dplyr_1.1.4 purrr_1.0.2
[7] readr_2.1.5 tidyr_1.3.0 tibble_3.2.1
[10] ggplot2_3.4.4 tidyverse_2.0.0 DiagrammeR_1.0.10
loaded via a namespace (and not attached):
[1] sass_0.4.8 utf8_1.2.4 generics_0.1.3 stringi_1.8.3
[5] hms_1.1.3 digest_0.6.34 magrittr_2.0.3 timechange_0.3.0
[9] evaluate_0.23 grid_4.3.2 RColorBrewer_1.1-3 fastmap_1.1.1
[13] rprojroot_2.0.4 workflowr_1.7.1 jsonlite_1.8.8 whisker_0.4.1
[17] promises_1.2.1 fansi_1.0.6 scales_1.3.0 jquerylib_0.1.4
[21] cli_3.6.2 rlang_1.1.3 visNetwork_2.1.2 ellipsis_0.3.2
[25] munsell_0.5.0 withr_3.0.0 cachem_1.0.8 yaml_2.3.8
[29] tools_4.3.2 tzdb_0.4.0 colorspace_2.1-0 httpuv_1.6.13
[33] vctrs_0.6.5 R6_2.5.1 lifecycle_1.0.4 git2r_0.33.0
[37] fs_1.6.3 htmlwidgets_1.6.4 pkgconfig_2.0.3 pillar_1.9.0
[41] bslib_0.6.1 later_1.3.2 gtable_0.3.4 glue_1.7.0
[45] Rcpp_1.0.12 highr_0.10 xfun_0.41 tidyselect_1.2.0
[49] rstudioapi_0.15.0 knitr_1.45 farver_2.1.1 htmltools_0.5.7
[53] labeling_0.4.3 rmarkdown_2.25 compiler_4.3.2