Linear regression examples
Environmental Data Analysis, Spring 2024
3/28/24
Goals
The examples below are meant to show you how to implement the three options for regression when you are violating the normality and/or equal variance assumptions:
- Remove outliers
- Transform the data
- Use a non-parametric model (Theil-Sen regression)
Example 1: Removing outliers
This dataset was generated by recording the miles traveled and kwh consumed each day, and recording the average temperature for the day from a local weather service. The question guiding our example is: How does ambient temperature affect an electric car’s efficiency + range?
library(tidyverse)
library(MASS)
library(mblm)
ecar <- read.csv("./data/ecar-and-temp-clean.csv")
str(ecar)'data.frame': 54 obs. of 3 variables:
$ date : chr "11/1/21" "11/3/21" "11/4/21" "11/6/21" ...
$ mi_kwh: num 3.42 3.52 3.28 2.91 2.87 ...
$ temp_F: num 58.4 55 51.4 48 47.2 48.2 54.4 51.2 57.6 55.8 ...Examine scatterplot
# Use ggplot to create a scatterplot to visualize the relationship between the two variables, putting the independent variable on the x axis and the dependent on the y axis
ggplot(data = ecar, aes(x = temp_F, y = mi_kwh)) +
geom_point() +
theme_bw() +
labs(x = "Temperature (deg F)", y = "Miles per kwh charge")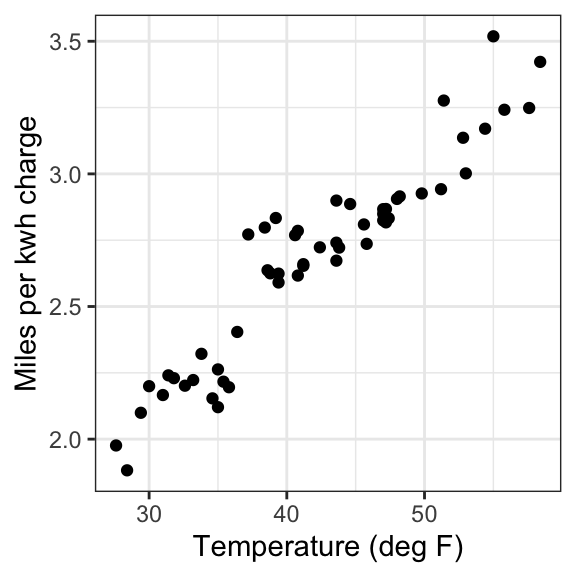
Fit a linear regression model + assess assumptions
Use the function lm() to fit a linear regression model
of mi/kwh as a function of temperature. The function requires the
following arguments:
- formula: a description of the model to be fitted (form: y ~ x)
- data: the name of the data frame containing the variables in the formula
mod <- lm(mi_kwh ~ temp_F, data = ecar)# Let's check the fit using diagnostic plots...
par(mfrow=c(2,2))
plot(mod)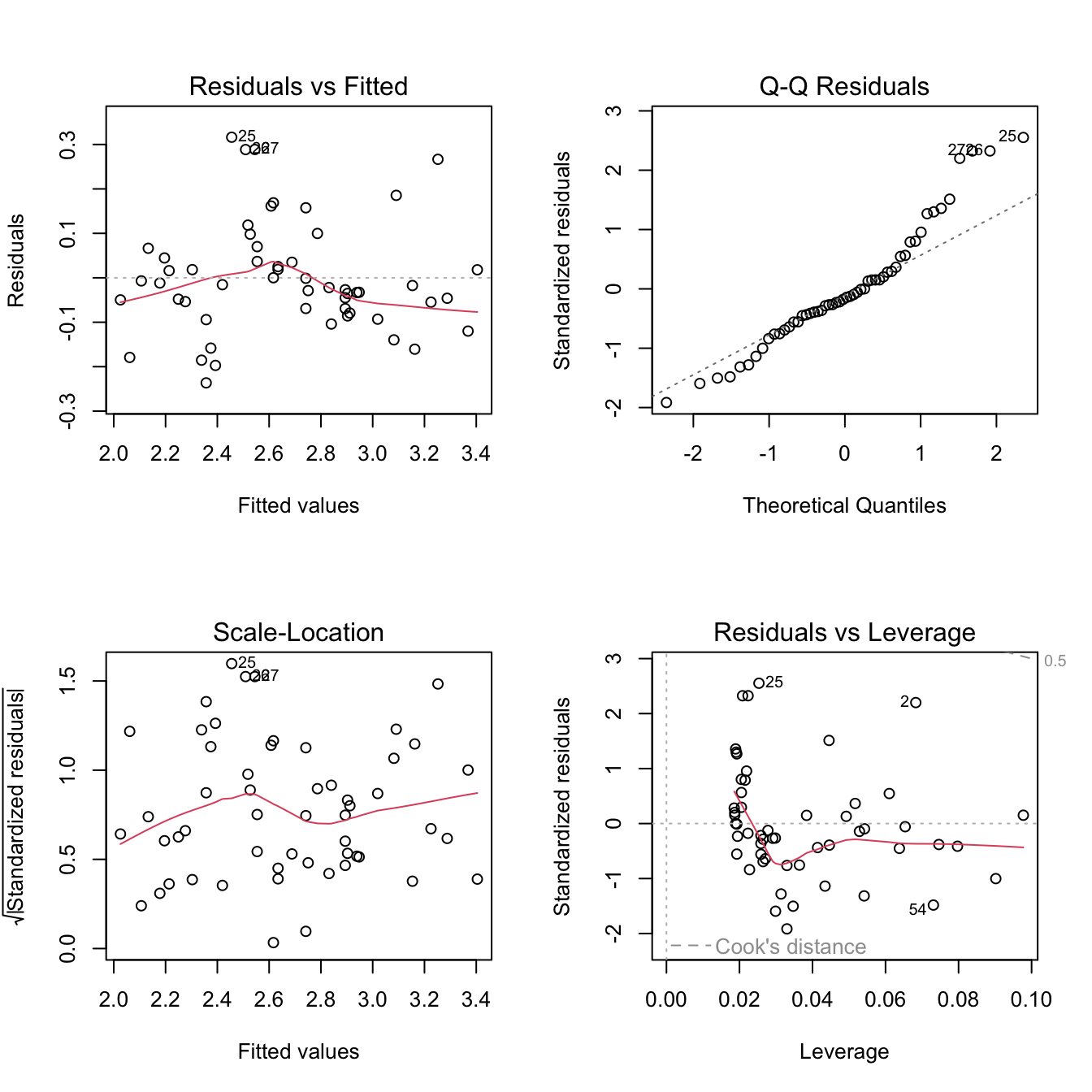
# And a histogram of the residuals...
hist(mod$residuals)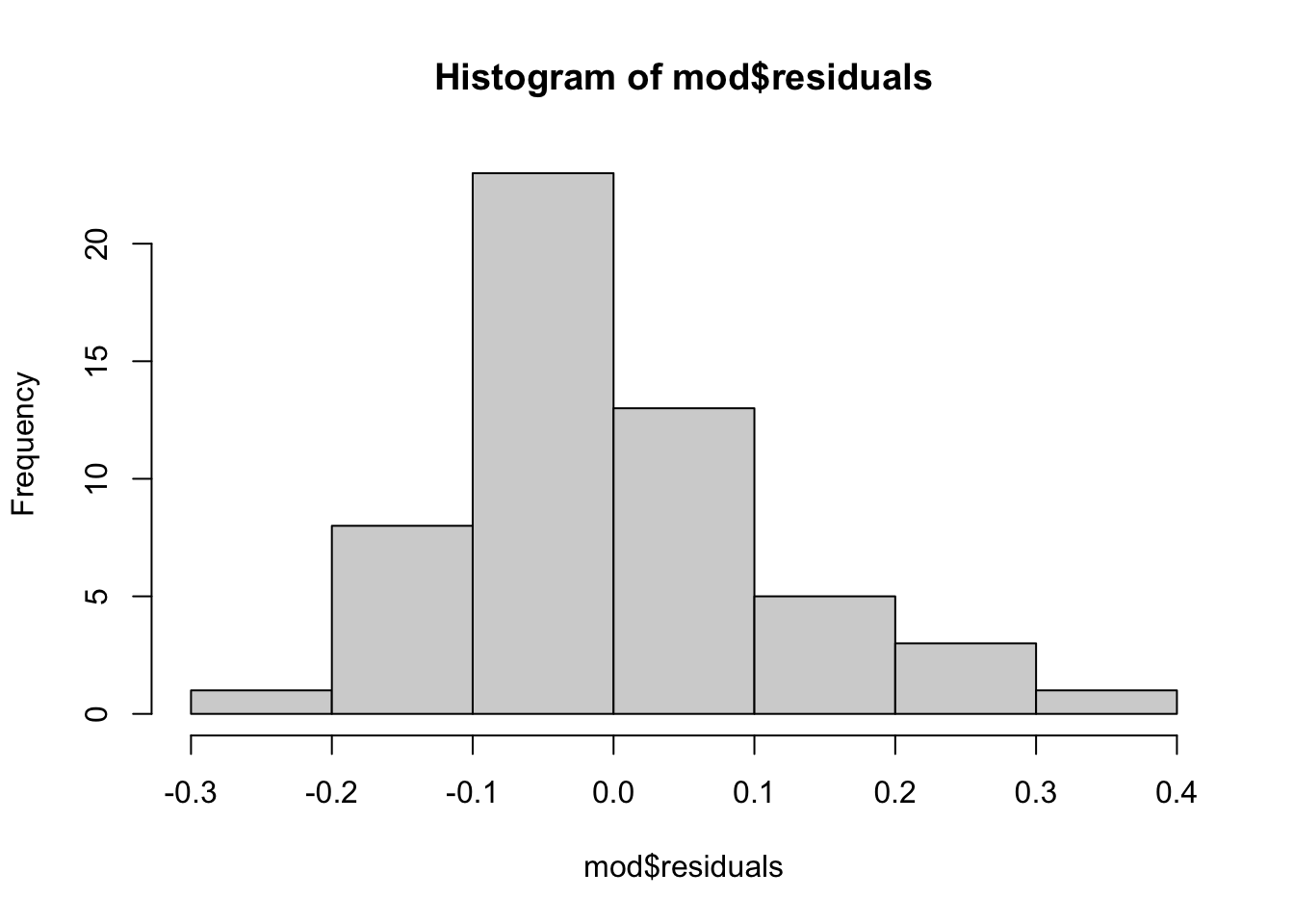
# Check mean and median of residuals
# Mean will always be zero, median may not be if not normal...
# We can see that the median is slightly below zero...
round(mean(mod$residuals), digits = 2)[1] 0round(median(mod$residuals), digits = 2)[1] -0.02# Create a new dataframe with the outliers marked
# First create a variable to store the row numbers
# Then use mutate + ifelse to mark the outliers
ecar_mark <- ecar %>%
mutate(rownum = seq(from = 1, to = 54, by = 1),
col = ifelse(rownum %in% c(25, 26, 27), "outlier", "not outlier"))
# Plot again with outliers marked
ggplot(data = ecar_mark, aes(x = temp_F, y = mi_kwh, colour = col)) +
geom_point() +
theme_bw() +
labs(x = "Temperature (deg F)", y = "Miles per kwh charge") +
theme(legend.title=element_blank(), legend.position = "top") +
scale_colour_manual(values = c("black","blue"))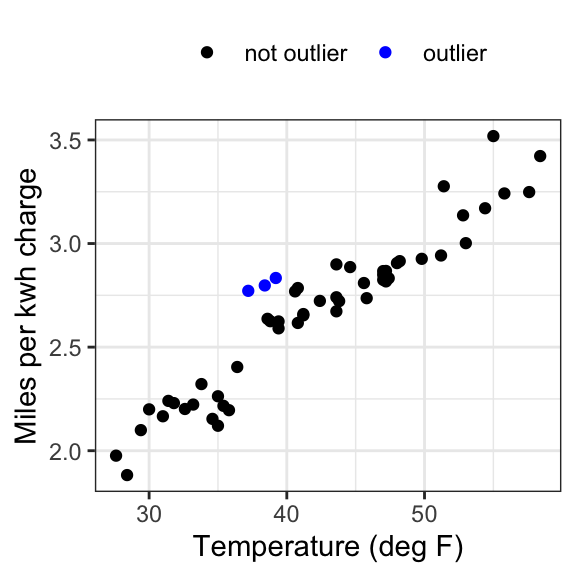
Exclude outliers + re-fit the model
We can see that there are a few potential outliers in our dataset (rows 25, 26, 27 - frequently marked as such on the diagnostic plots). These do not look too extreme, but if we are worried about them, we can refit the model without those points to see if it changes the outcome.
# To remove those rows from the data we will use a skill called 'indexing'
# The negative sign indicates we want to remove rows 25, 26, 27
# The comma after the parenthesis is important -
# leaving the second part blank selects all columns
ecar_no_outliers <- ecar[-c(25,26,27), ]
# Now let's refit the model...
mod_no_outliers <- lm(mi_kwh ~ temp_F, data = ecar_no_outliers)# Since we created a column, we can easily filter
ecar_no_outliers <- ecar_mark %>%
filter(col != "outlier") # notice the exclamation point! (means 'not')
# Now let's refit the model...
mod_no_outliers <- lm(mi_kwh ~ temp_F, data = ecar_no_outliers)# And check the fit...
par(mfrow=c(2,2))
plot(mod_no_outliers)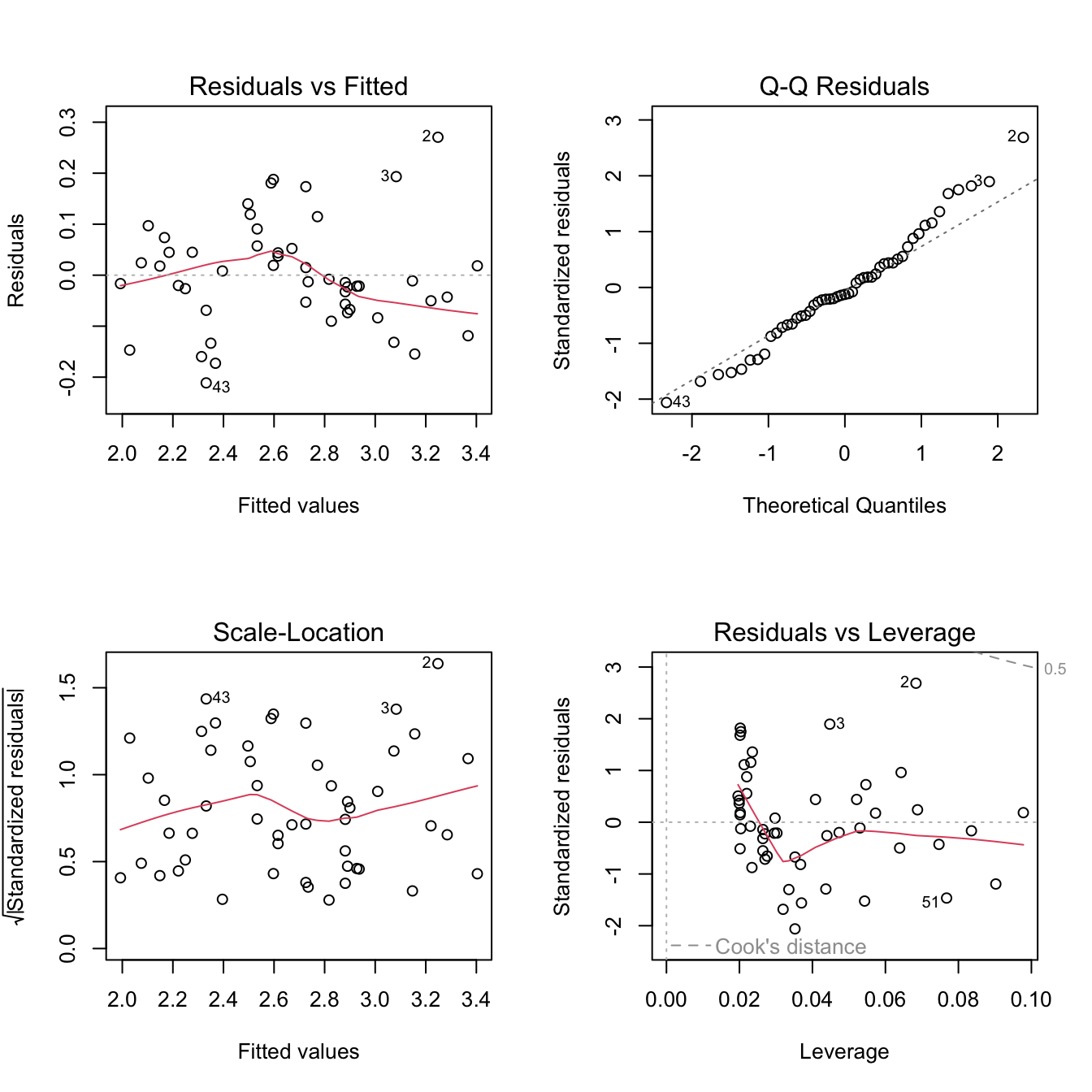
hist(mod_no_outliers$resid)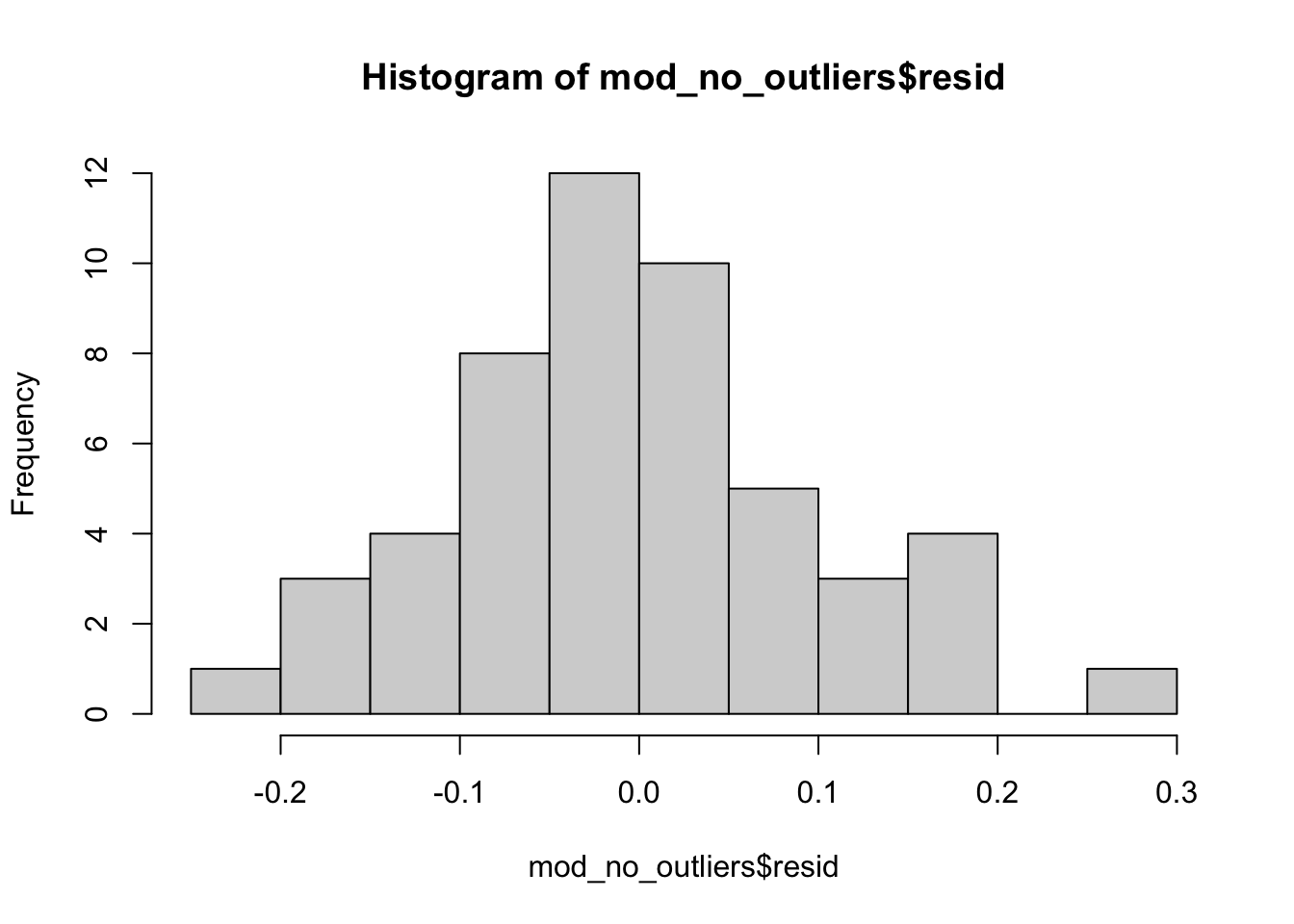
Compare models
Do your conclusions change?
summary(mod)
Call:
lm(formula = mi_kwh ~ temp_F, data = ecar)
Residuals:
Min 1Q Median 3Q Max
-0.23648 -0.06920 -0.01967 0.04267 0.31633
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 0.790601 0.092151 8.579 1.56e-11 ***
temp_F 0.044753 0.002156 20.761 < 2e-16 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 0.1255 on 52 degrees of freedom
Multiple R-squared: 0.8923, Adjusted R-squared: 0.8903
F-statistic: 431 on 1 and 52 DF, p-value: < 2.2e-16summary(mod_no_outliers)
Call:
lm(formula = mi_kwh ~ temp_F, data = ecar_no_outliers)
Residuals:
Min 1Q Median 3Q Max
-0.21124 -0.06210 -0.01290 0.04861 0.27054
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 0.728025 0.077560 9.387 1.58e-12 ***
temp_F 0.045820 0.001804 25.401 < 2e-16 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 0.1043 on 49 degrees of freedom
Multiple R-squared: 0.9294, Adjusted R-squared: 0.928
F-statistic: 645.2 on 1 and 49 DF, p-value: < 2.2e-16Example 2: Transformation
In the previous example, our data reasonably fit the assumptions. Now let’s look at an example where that is not the case and see how we might adjust the data to meet assumptions.
We will use a dataset called mammals that contains
average brain weights (in g) and body weights (in kg) for 62 species of
land mammals. We want to know if mammals with heavier bodies also have
heavier brains.
Preliminary plots
# Use ggplot to examine the relationship between body weight and brain weight
ggplot(data = mammals, aes(x = body, y = brain)) +
geom_point(alpha = .5) +
theme_bw() +
labs(x = "Body weight (kg)", y = "Brain weight (g)")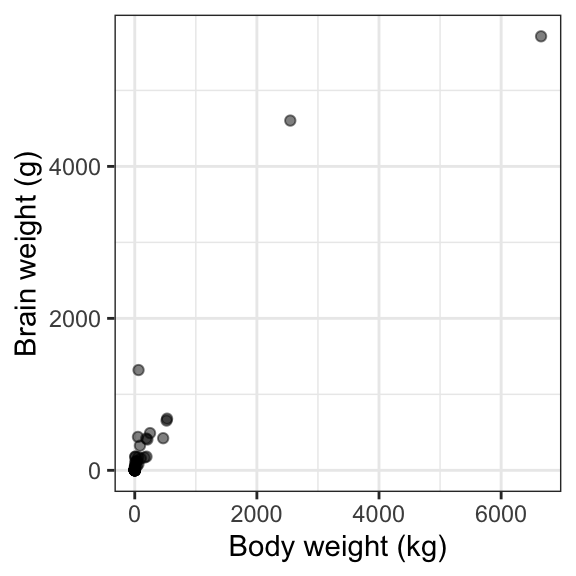
# Use ggplot to examine the histograms for each variable
ggplot(data = mammals, aes(x = body)) +
geom_density() +
theme_bw()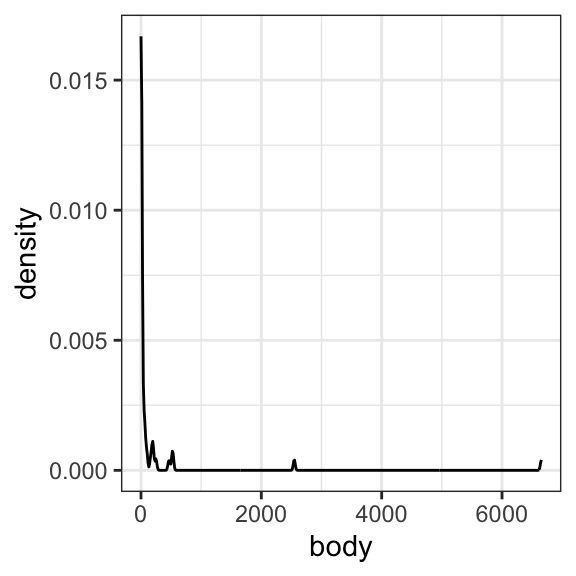
ggplot(data = mammals, aes(x = brain)) +
geom_density() +
theme_bw()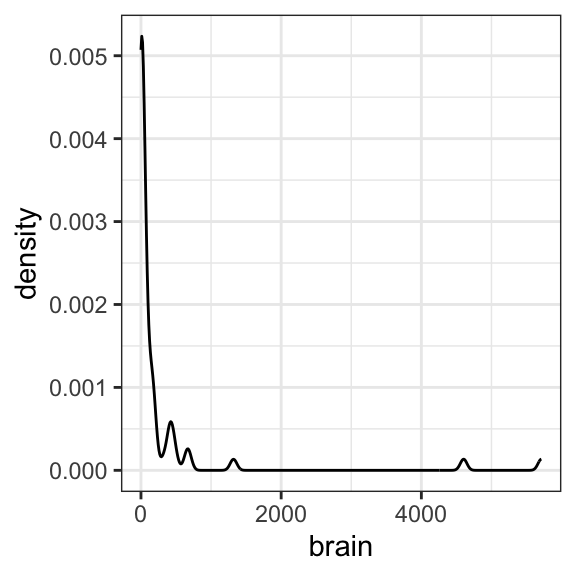
Fit a linear regression model + assess assumptions
Now let’s see what happens when we fit a linear model to these data.
mod_raw <- lm(brain ~ body, data = mammals)par(mfrow=c(2,2))
plot(mod_raw)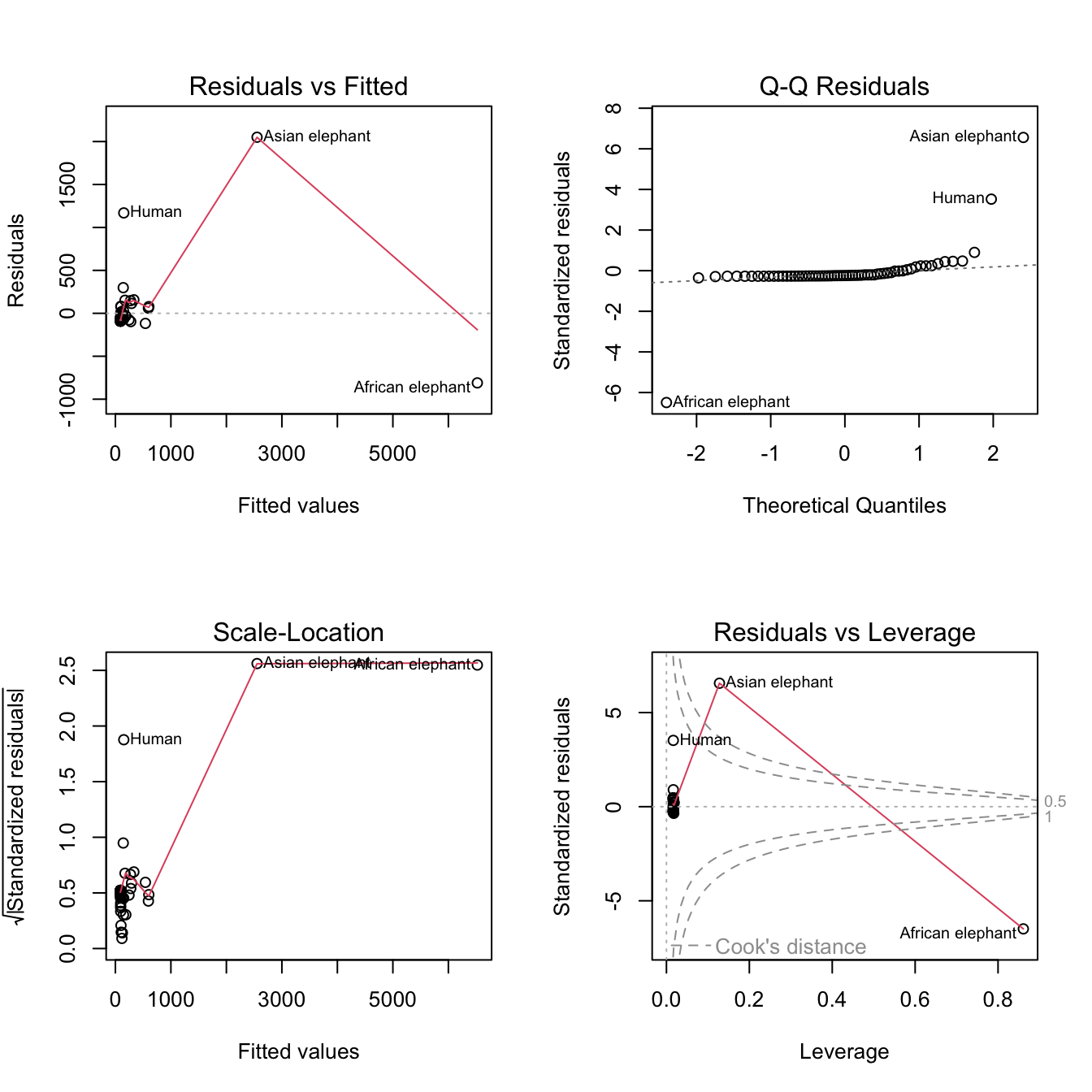
hist(mod_raw$residuals)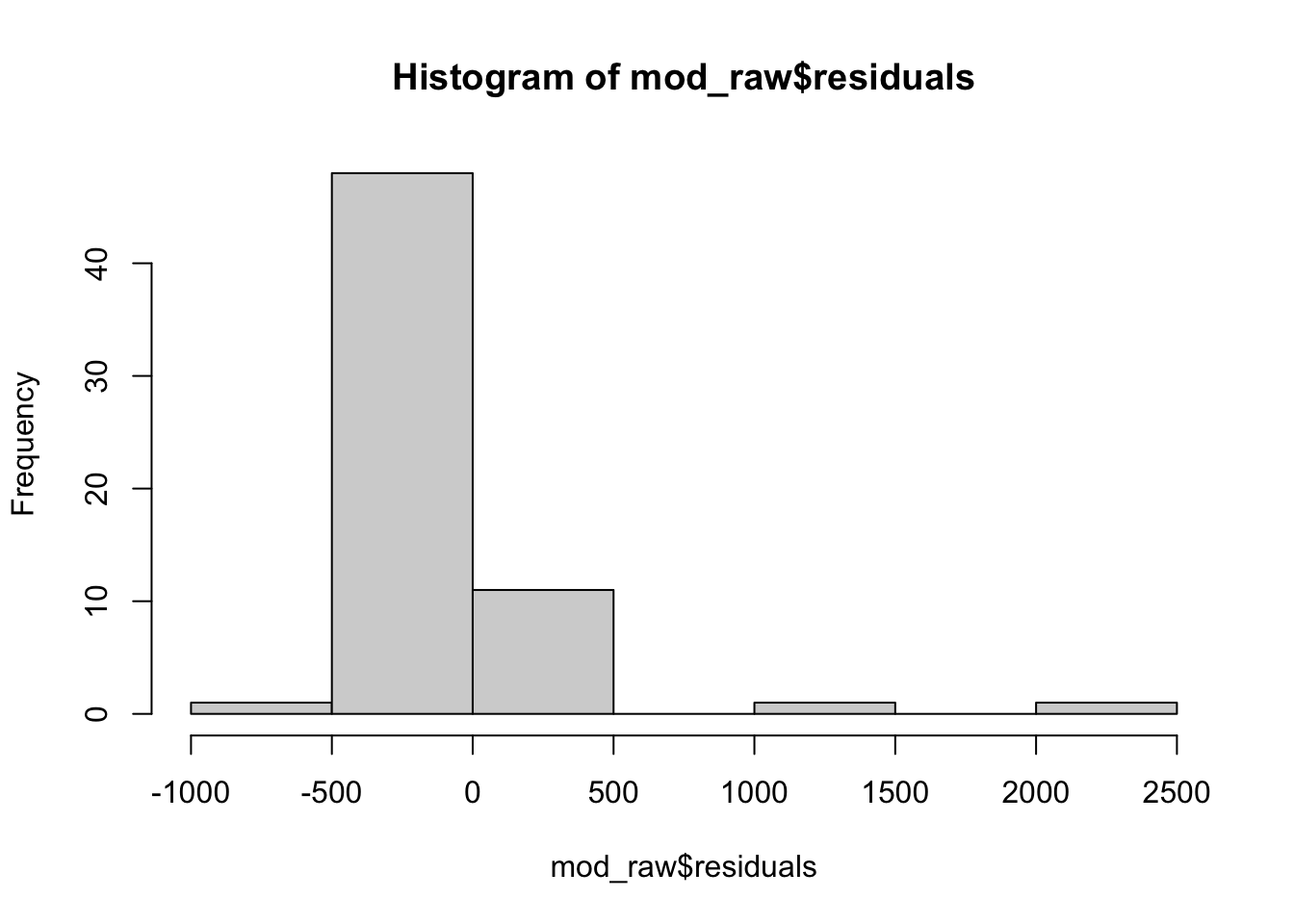
Let’s see what happens when we examine the model output:
summary(mod_raw)
Call:
lm(formula = brain ~ body, data = mammals)
Residuals:
Min 1Q Median 3Q Max
-810.07 -88.52 -79.64 -13.02 2050.33
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 91.00440 43.55258 2.09 0.0409 *
body 0.96650 0.04766 20.28 <2e-16 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 334.7 on 60 degrees of freedom
Multiple R-squared: 0.8727, Adjusted R-squared: 0.8705
F-statistic: 411.2 on 1 and 60 DF, p-value: < 2.2e-16Transform the data + re-fit the model
# Use ggplot to examine the relationship between body weight and brain weight
ggplot(data = mammals, aes(x = log10(body), y = log10(brain))) +
geom_point() +
theme_bw()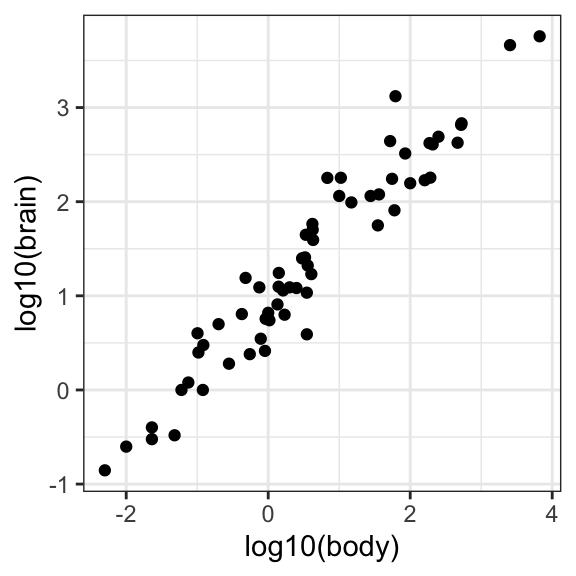
# Use ggplot to examine the histograms for each variable
ggplot(data = mammals, aes(x = log10(body))) +
geom_density() +
theme_bw()
ggplot(data = mammals, aes(x = log10(brain))) +
geom_density() +
theme_bw()
This looks promising, now let’s fit a new model with the transformed variables…
# Fit a new model using log10() to transform both variables
mod_log10 <- lm(log10(brain) ~ log10(body), data = mammals)par(mfrow=c(2,2))
plot(mod_log10)
hist(mod_log10$residuals)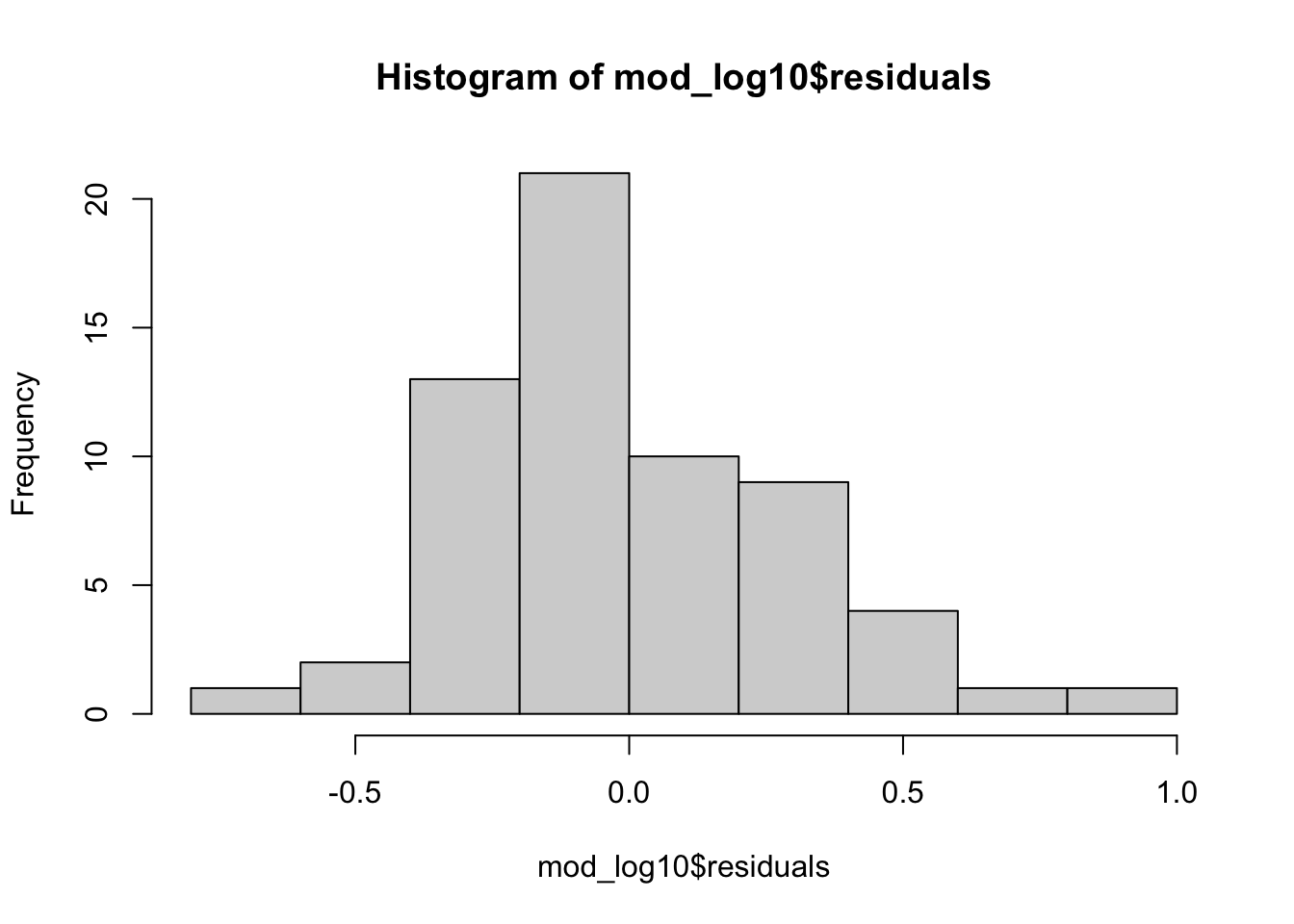
Compare the two models
Let’s compare the model output from the model on the raw data and the model on transformed data:
How would your interpretation of the second model differ from the first? (hint: consider the units of the slope and intercept)
# Use `summary` to extract the output from the two models
summary(mod_raw)
Call:
lm(formula = brain ~ body, data = mammals)
Residuals:
Min 1Q Median 3Q Max
-810.07 -88.52 -79.64 -13.02 2050.33
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 91.00440 43.55258 2.09 0.0409 *
body 0.96650 0.04766 20.28 <2e-16 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 334.7 on 60 degrees of freedom
Multiple R-squared: 0.8727, Adjusted R-squared: 0.8705
F-statistic: 411.2 on 1 and 60 DF, p-value: < 2.2e-16summary(mod_log10)
Call:
lm(formula = log10(brain) ~ log10(body), data = mammals)
Residuals:
Min 1Q Median 3Q Max
-0.74503 -0.21380 -0.02676 0.18934 0.84613
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 0.92713 0.04171 22.23 <2e-16 ***
log10(body) 0.75169 0.02846 26.41 <2e-16 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 0.3015 on 60 degrees of freedom
Multiple R-squared: 0.9208, Adjusted R-squared: 0.9195
F-statistic: 697.4 on 1 and 60 DF, p-value: < 2.2e-16Graph results
Finally, let’s create a graph to present the results of our final model. This time we will include the error band around the slope.
ggplot(data = mammals, aes(x = log10(body), y = log10(brain))) +
geom_point() +
geom_smooth(method = 'lm', se = TRUE) +
labs(x = "log10 - Body weight (kg)", y = "log10 - Brain weight (g)") +
theme_bw()`geom_smooth()` using formula = 'y ~ x'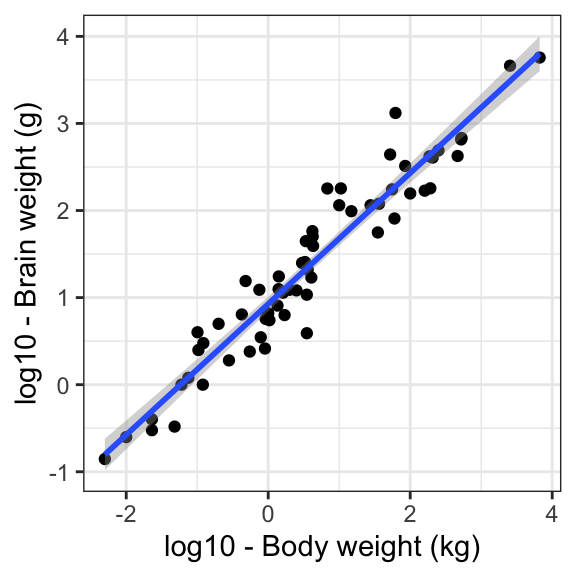
Example 3: Theil-Sen regression
Now let’s look at an example where assumptions are not so clearly met and instead of transformation we take a different approach to the model all together - using a non-parametric alternative called Theil-Sen Regression. We will use the dataset from Tuesday’s lab (Garvin et al. 2006), but look at a slightly different question:
How is does zinc content differ depending on year of variety release?
Predictor (Independent):
yearResponse (Dependent):
seed_Zn_ug_g
garvin <- read.csv("./data/Garvin_2006_data.csv")
str(garvin)'data.frame': 14 obs. of 3 variables:
$ year : int 1919 1942 1944 1964 1967 1970 1978 1982 1984 1992 ...
$ yield_kg_ha : num 3022 3066 3261 4016 3365 ...
$ seed_Zn_ug_g: num 26.3 25.5 21.7 22.6 23 ...Preliminary graphs
# CODE: Use ggplot to examine the relationship between year of release and zinc
ggplot(data = garvin, aes(x = year, y = seed_Zn_ug_g)) +
geom_point() +
geom_smooth(method = 'loess', se = FALSE) +
theme_bw()`geom_smooth()` using formula = 'y ~ x'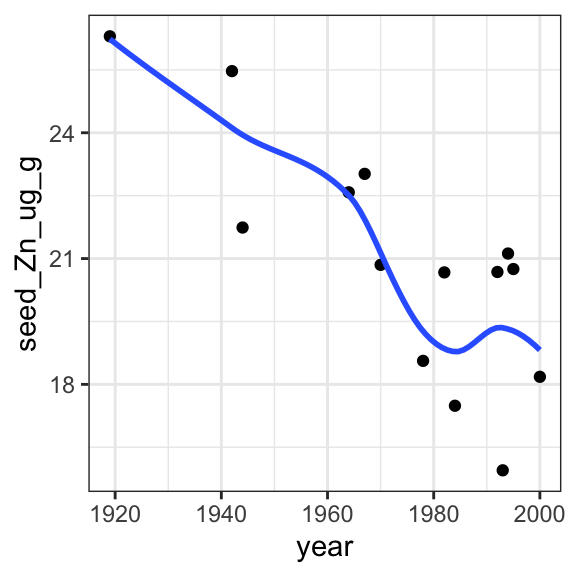
# CODE: Use ggplot to examine the histograms for each variable
ggplot(data = garvin, aes(x = year)) +
geom_density() +
theme_bw()
ggplot(data = garvin, aes(x = seed_Zn_ug_g)) +
geom_density() +
theme_bw()
Fit the linear regression model + assess assumptions
Now let’s see what happens when we fit a linear model to these data.
# CODE: Use 'lm' to fit a linear regression model of zinc content as a function of year
mod_raw <- lm(seed_Zn_ug_g ~ year, data = garvin)# CODE: Use 'plot' and 'hist' to generate diagnostic plots of the residuals from the model
par(mfrow = c(2,2))
plot(mod_raw)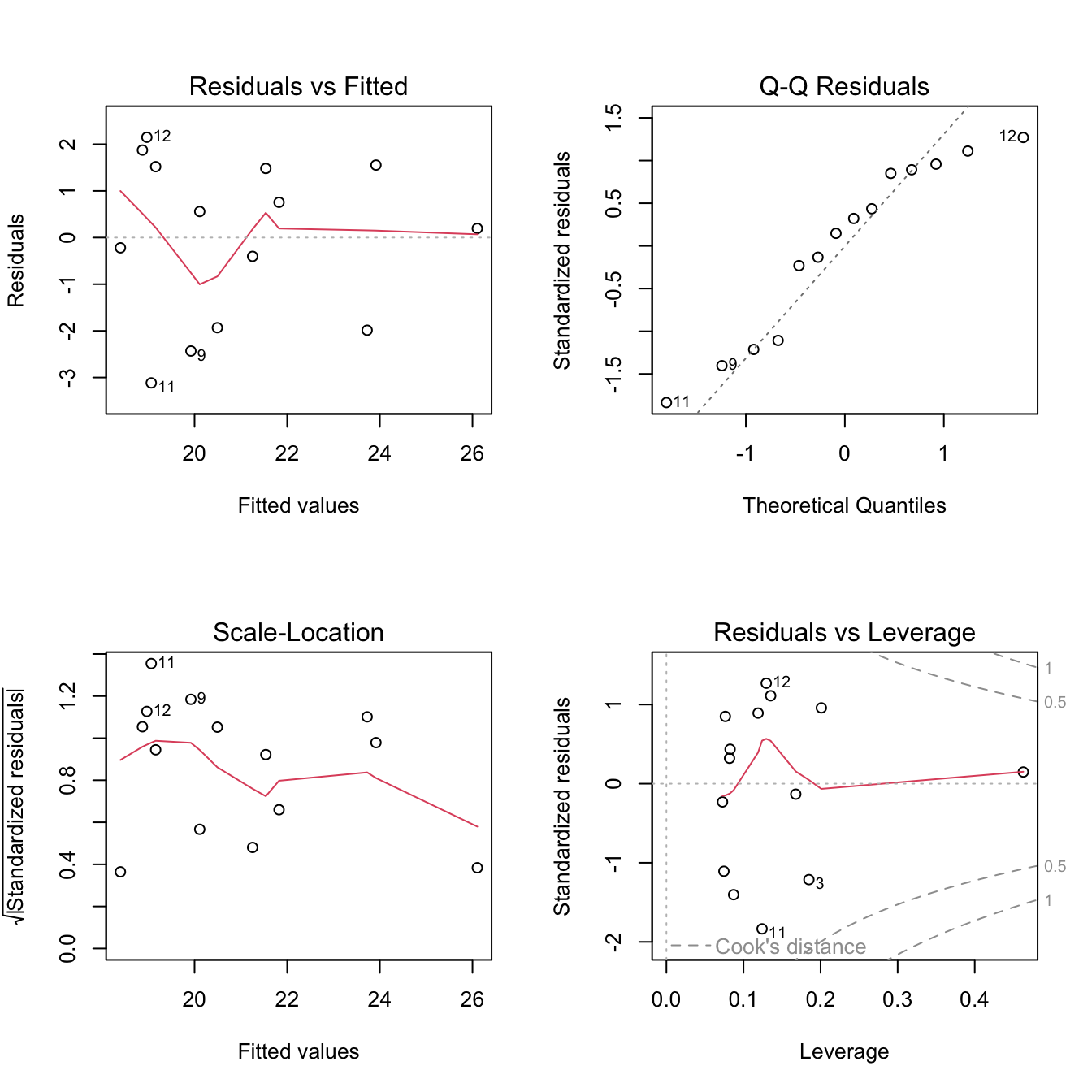
hist(mod_raw$resid)
Use Theil-Sen regression to fit an alternative model
# Fit a new model using function `mblm` (median-based linear models)
mod_med <- mblm(seed_Zn_ug_g ~ year, data = garvin)
# We do not look at the distribution of residuals because this method does not make assumptions about the distribution of the residualsCompare the two models
# Use `summary` to extract the output from the two models
summary(mod_raw)
Call:
lm(formula = seed_Zn_ug_g ~ year, data = garvin)
Residuals:
Min 1Q Median 3Q Max
-3.116 -1.550 0.377 1.510 2.150
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 208.63105 41.36964 5.043 0.000288 ***
year -0.09512 0.02096 -4.537 0.000682 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 1.814 on 12 degrees of freedom
Multiple R-squared: 0.6317, Adjusted R-squared: 0.601
F-statistic: 20.58 on 1 and 12 DF, p-value: 0.0006815summary(mod_med)
Call:
mblm(formula = seed_Zn_ug_g ~ year, dataframe = garvin)
Residuals:
Min 1Q Median 3Q Max
-4.0374 -2.5898 -0.7665 0.3538 1.2347
Coefficients:
Estimate MAD V value Pr(>|V|)
(Intercept) 223.38411 74.75111 105 0.00109 **
year -0.10206 0.03911 0 0.00109 **
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 2.152 on 12 degrees of freedomconfint(mod_med, level = 0.95) 0.025 0.975
(Intercept) 140.2280043 273.80303768
year -0.1283798 -0.06004539Graph results
Theil-Sen regression does not have a built in function in geom_smooth, so we’ll need to take a different approach to fitting our trendline
# To see how the coef() function works, check this out:
coef(mod_med)(Intercept) year
223.3841111 -0.1020556 # Rather than use geom_smooth, we will use geom_abline (use help file to see argument!)
# the function coef() extracts coefficients from the model
ggplot(data = garvin, aes(x = year, y = seed_Zn_ug_g)) +
geom_point() +
geom_abline(intercept = coef(mod_med)[1], slope = coef(mod_med)[2], color = "blue") +
labs(x = "Year of variety introduction", y = "Zinc content (ug/g)") +
theme_light() 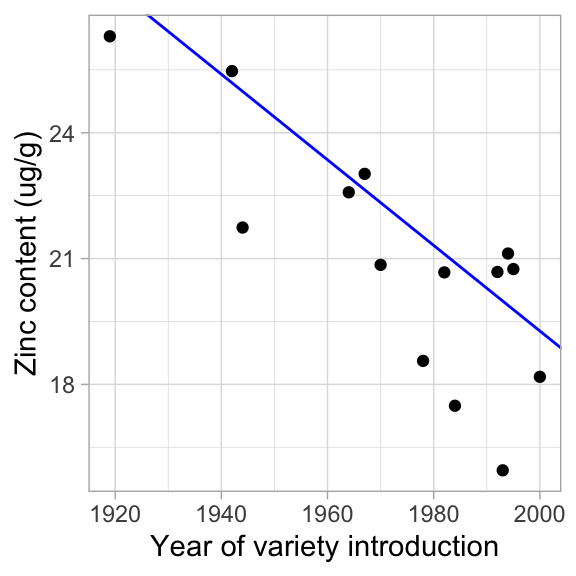 How similar is it to the OLS linear regression line?
How similar is it to the OLS linear regression line?
# Here is the graph with the standard regression line
ggplot(data = garvin, aes(x = year, y = seed_Zn_ug_g)) +
geom_point() +
geom_smooth(method = "lm") +
labs(x = "Year of variety introduction", y = "Zinc content (ug/g)") +
theme_light() `geom_smooth()` using formula = 'y ~ x'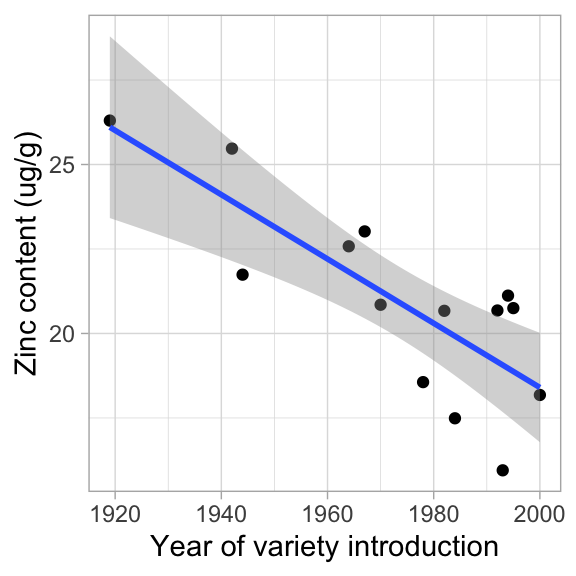
sessionInfo()R version 4.3.2 (2023-10-31)
Platform: x86_64-apple-darwin20 (64-bit)
Running under: macOS Monterey 12.4
Matrix products: default
BLAS: /Library/Frameworks/R.framework/Versions/4.3-x86_64/Resources/lib/libRblas.0.dylib
LAPACK: /Library/Frameworks/R.framework/Versions/4.3-x86_64/Resources/lib/libRlapack.dylib; LAPACK version 3.11.0
locale:
[1] en_US.UTF-8/en_US.UTF-8/en_US.UTF-8/C/en_US.UTF-8/en_US.UTF-8
time zone: America/New_York
tzcode source: internal
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] mblm_0.12.1 MASS_7.3-60 lubridate_1.9.3 forcats_1.0.0
[5] stringr_1.5.1 dplyr_1.1.4 purrr_1.0.2 readr_2.1.5
[9] tidyr_1.3.0 tibble_3.2.1 ggplot2_3.4.4 tidyverse_2.0.0
loaded via a namespace (and not attached):
[1] sass_0.4.8 utf8_1.2.4 generics_0.1.3 lattice_0.21-9
[5] stringi_1.8.3 hms_1.1.3 digest_0.6.34 magrittr_2.0.3
[9] timechange_0.3.0 evaluate_0.23 grid_4.3.2 fastmap_1.1.1
[13] Matrix_1.6-1.1 rprojroot_2.0.4 workflowr_1.7.1 jsonlite_1.8.8
[17] promises_1.2.1 mgcv_1.9-0 fansi_1.0.6 scales_1.3.0
[21] jquerylib_0.1.4 cli_3.6.2 rlang_1.1.3 splines_4.3.2
[25] munsell_0.5.0 withr_3.0.0 cachem_1.0.8 yaml_2.3.8
[29] tools_4.3.2 tzdb_0.4.0 colorspace_2.1-0 httpuv_1.6.13
[33] vctrs_0.6.5 R6_2.5.1 lifecycle_1.0.4 git2r_0.33.0
[37] fs_1.6.3 pkgconfig_2.0.3 pillar_1.9.0 bslib_0.6.1
[41] later_1.3.2 gtable_0.3.4 glue_1.7.0 Rcpp_1.0.12
[45] highr_0.10 xfun_0.41 tidyselect_1.2.0 rstudioapi_0.15.0
[49] knitr_1.45 farver_2.1.1 nlme_3.1-163 htmltools_0.5.7
[53] labeling_0.4.3 rmarkdown_2.25 compiler_4.3.2